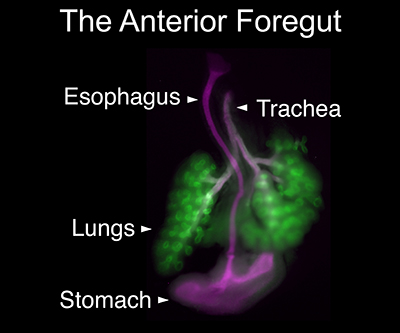
Microscopy image of the anterior foregut.
Credit: Rocha Lab, NICHD
Genes are often visualized as long strings that are highly organized within the cell’s nucleus, with mechanisms that help fold, compact, and unfold areas as needed to direct cellular growth, function, and development.
DNA loops created by the CTCF protein are one type of regulatory mechanism. These loops create physical boundaries between enhancers—non-coding DNA that helps promote gene activity or transcription—and the areas of genes where transcription begins, which are known as promoters. For unclear reasons, CTFC can act strongly to repress gene activity in certain types of tissues and not in others.
In a recent mouse study by the Rocha Lab, researchers explored this phenomenon by defining the contexts that allow some enhancer-promoter interactions to bypass CTCF-mediated loops. They found that CTCF loops can interfere with expression of the Sox2 gene, which is essential for embryonic development, in the anterior foregut—the precursor of the lungs, trachea, esophagus and stomach. However, the team also observed that CTCF did not repress Sox2 activation in epiblast and neural tissues. The authors propose that enhancer clusters with a high density of regulatory activity can better overcome physical barriers to maintain faithful gene expression and phenotypic robustness. This work is also featured in the NIH Catalyst.
Learn more about the Genetics and Epigenetics of Development Affinity Group:
https://www.nichd.nih.gov/about/org/dir/affinity-groups/GED.
 BACK TO TOP
BACK TO TOP